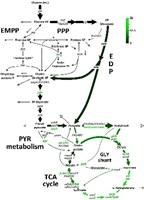
 Pseudomonas aeruginosa can be a dangerous bacterium. It is one of the main causes of serious hospital-acquired infections and it is also a leading cause of death among people with cystic fibrosis. Researchers are studying the bacterium to find out what makes it so dangerous - an important step if we are to develop better ways to protect people from this deadly infection. Here, we use a systems biology approach to understand the contribution of the core metabolic machinery of P. aeruginosa to its high robustness and resistance. We study laboratory strains and clinical isolates from urinary tract infections on the level of metabolic fluxes. Particularly, we are interested to understand, how the pathogen withstands oxidative stress, imposed by the host defence. The selected isolates are genetically diverse, exhibit heterogeneous production of virulence factors in vitro and display an excellent collection to search for conserved metabolic strategies and niche specific adaptations in this important human pathogen. We cooperate with the groups of Dieter Jahn (Microbiology, Technische Universität Braunschweig) and Kathrin Riedel (University of Greifswald).
Pseudomonas aeruginosa can be a dangerous bacterium. It is one of the main causes of serious hospital-acquired infections and it is also a leading cause of death among people with cystic fibrosis. Researchers are studying the bacterium to find out what makes it so dangerous - an important step if we are to develop better ways to protect people from this deadly infection. Here, we use a systems biology approach to understand the contribution of the core metabolic machinery of P. aeruginosa to its high robustness and resistance. We study laboratory strains and clinical isolates from urinary tract infections on the level of metabolic fluxes. Particularly, we are interested to understand, how the pathogen withstands oxidative stress, imposed by the host defence. The selected isolates are genetically diverse, exhibit heterogeneous production of virulence factors in vitro and display an excellent collection to search for conserved metabolic strategies and niche specific adaptations in this important human pathogen. We cooperate with the groups of Dieter Jahn (Microbiology, Technische Universität Braunschweig) and Kathrin Riedel (University of Greifswald).
- Systems biology of various P. aeruginosa isolates
- Systems-wide 13C metabolic flux analysis
- Integration of metabolome, proteome and transcriptome data
- Elucidation of cellular processes during catheter-associated urinary tract infections
Publications
Dolan, S, Kohlstedt, M, Trigg, S, Ramirez, P, Kaminski, C, Wittmann, C, Welch, M (2020) Contextual flexibility in Pseudomonas aeruginosa central carbon metabolism during growth in single carbon sources. mBio 11:e02684-19. Link.
Kohlstedt, M, Wittmann, C (2019) GC-MS based 13C metabolic flux analysis resolves the cyclic glucose metabolism of Pseudomonas putida KT2440 and Pseudomonas aeruginosa PAO1. Metab. Eng. 54:35-53. Link
van Duuren, JBJH, Müsken, M, Karge, B, Tomasch, J, Wittmann, C, Häussler, S, Brönstrup, M (2017) Use of Single-Frequency impedance spectroscopy to characterize the growth dynamics of biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 7:5223. PUBMED
Lassek C, Berger A, Zühlke D, Wittmann C, Riedel K (2016) Proteome and carbon flux analysis of P. aeruginosa clinical isolates from different infection sites. Proteomics. 16:1381-1385. PUBMED
Berger A, Dohnt K, Tielen P, Jahn D, Becker J, Wittmann C (2014) Robustness and plasticity of metabolic pathway flux among uropathogenic isolates of Pseudomonas aeruginosa. PLoS ONE. 9(4):e88368. PUBMED
Partners

